Overview: The current version of cncRNAdb documents more than 2,600 manually curated entries of cncRNA functions with experimental evidence, involving more than 2,000 RNAs (including over 1,300 translated ncRNAs and over 600 untranslated mRNAs) across over 20 species. In summary, cncRNAdb provides a user-friendly interface to query, browse and visualize detailed information about these cncRNAs and will be of help in integrating, analyzing and predicting cncRNAs, enabling faster structural and functional research of RNA.
The homepage is displayed in the following Fig 1-1.
Fig 1-1:
1. Main functions of the database are provided in menu bar form (boxed in red).
2. Introduction and overview for cncRNAdb.
3. Data statistics of cncRNAdb.
4. Quick browse for users (by clicks the pictures of different types of cncRNA).
5. Other databases contributed by our group.
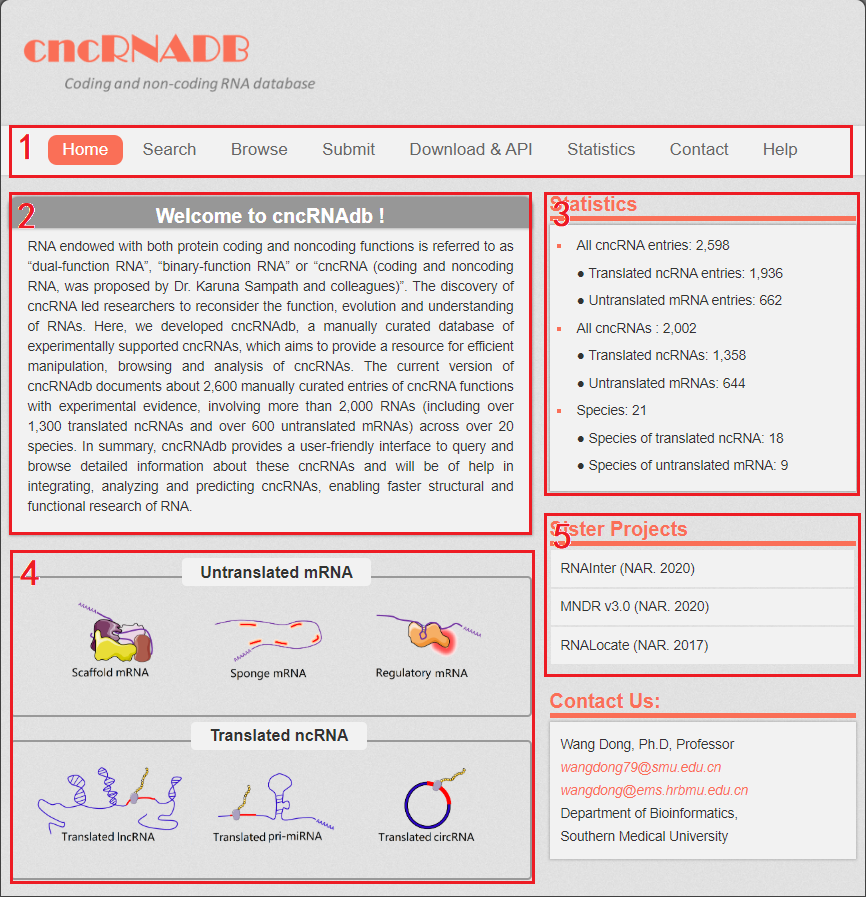
Fig 1-1 Homepage
The search page is displayed in Fig 2-1:
Keyword search: users can enter a keyword (RNA name or disease name) to search cncRNA.
1. Carefully select a dataset: Two choices are provided.
2. Select the species to do the search.
3. Select gene name or disease name to do search.
4. Enter a keyword corresponding to selected dataset.
5. Select if you want to exactly search the keyword, or we will search the keyword in a fuzzy pattern.
Locus search: users can enter a genome location to search cncRNA.
6. Carefully select a dataset: Two choices are provided.
7. Select the species to do the search.
8. Chose the right chromosome.
9. Enter the start and end of the genome location.
Blast search: users can enter a nucleotide/peptide sequence to search cncRNA.
10. Carefully select a dataset: Two choices are provided.
11. Select the sequence type.
12. Enter a sequence to search cncRNA.
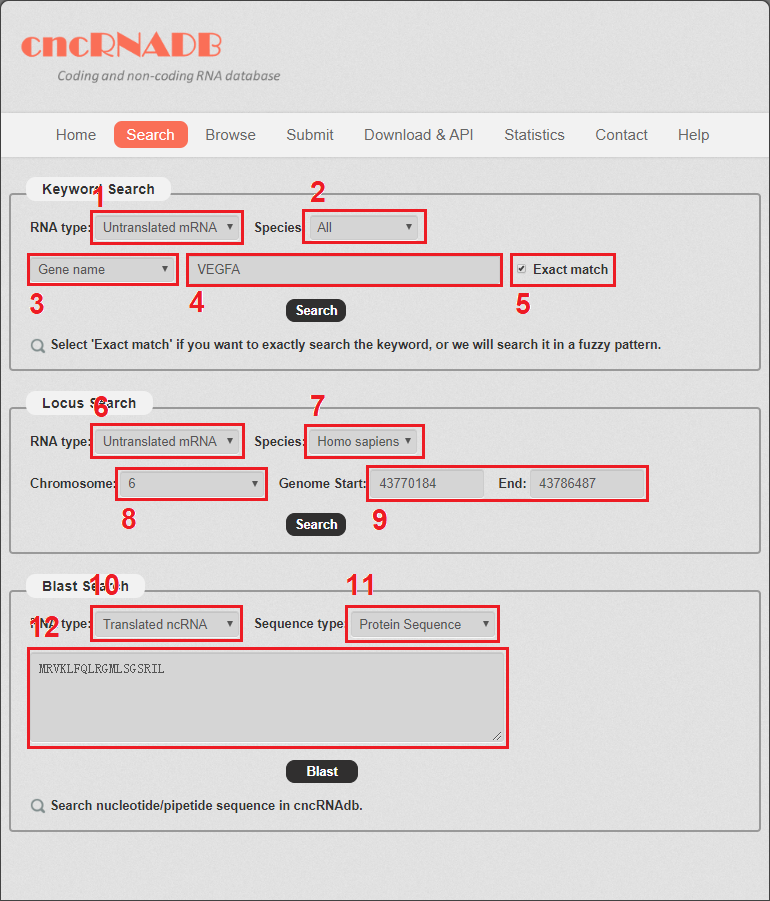
Fig 2-1 Search page
1. For keyword Search: First we have to choose the RNA type. There are two types in our search as the picture shows. In this example, we choose Translated ncRNA.
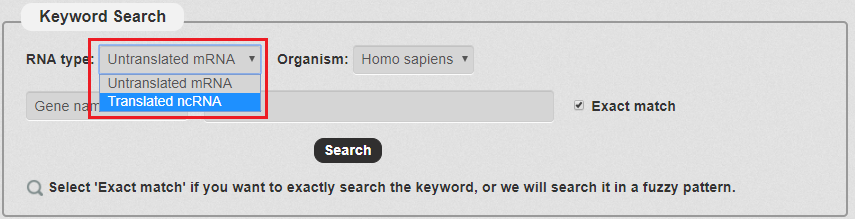
Fig 3-1
2. Next, we have to choose the Organism. There are some species in our search as the picture shows. In this example, we choose Medicago truncatula .
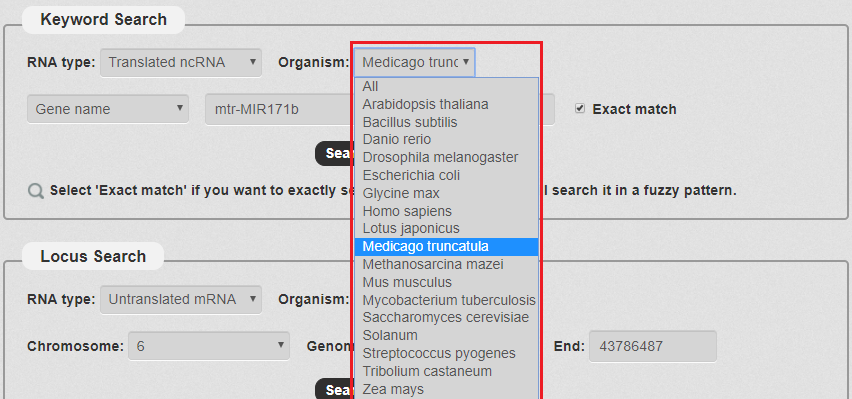
Fig 3-2
3. Then, we have to choose the keyword type. There are two types in our search as the picture shows. In this example, we choose Gene name.
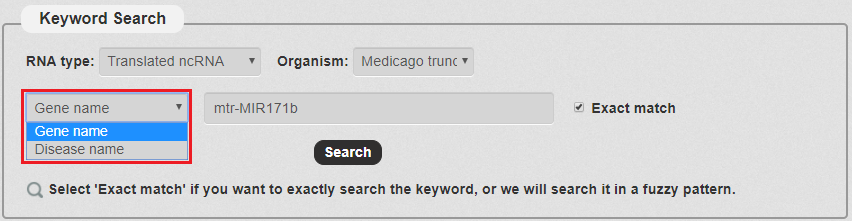
Fig 3-3
4. Then, we enter the keyword according to the keyword type selected in the previous step. In this example, we choose 'mtr-MIR171b' as the keyword.With all the filters, we can click 'Search' to query the result.
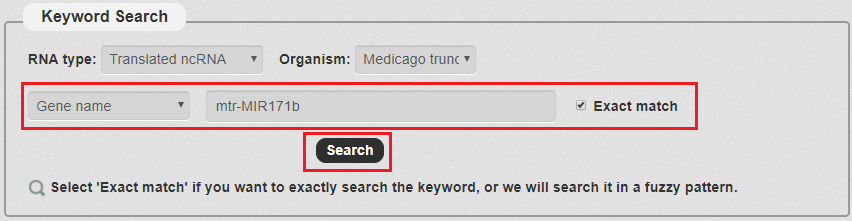
Fig 3-4
5. After few seconds, the result will occur. All the cncRNAs are represented in the table format, and your filters and the total numbers of entries are in the head of the web page.
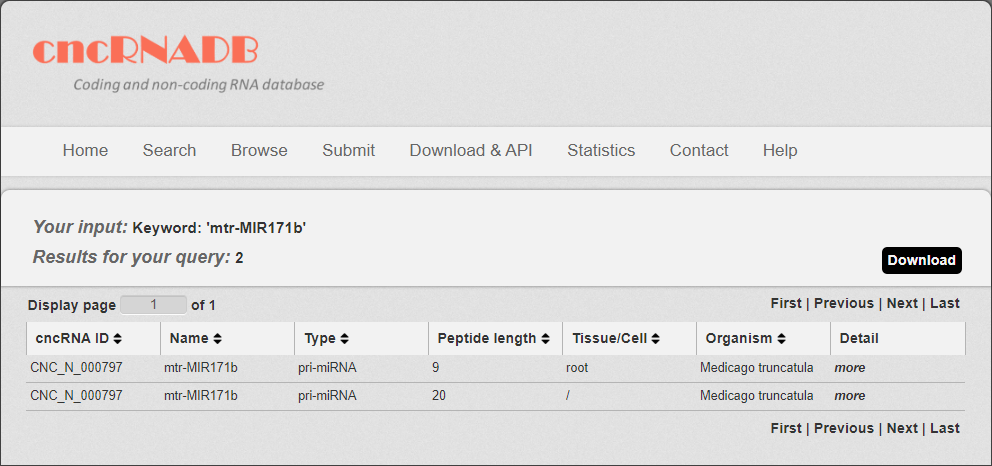
Fig 3-5
In the result page, all entries are listed with basic information including cncRNA ID, RNA symbols, cncRNA type, Peptide length (only for translated ncRNA), tissue and species.
Fig 3-1:
1. Your current input conditions.
2. Total sum of results.
3. Download your search results.
4. Click to turn the page.
5. The result table.
6. Click to link to Detail page.
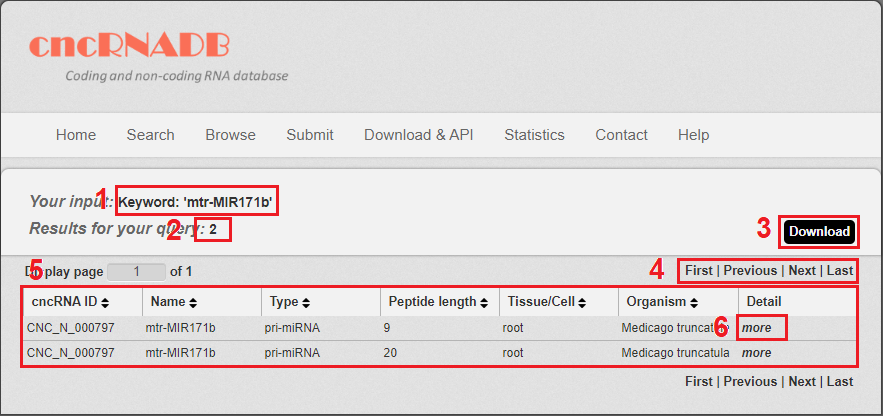
Fig 4-1 Result page
In the “Detail” page, you can get the detail information of the cncRNA including “Basic Information”, “Peptide information” (only for translated ncRNA), “Sequence and evidence support”, “References” and “related subcellular localization, related diseases and related interactions (from OMIM, MNDR v3.0, RNALocate and RNAInter database)”.
Fig 5-1:
1. Basic Information: including cncdb ID, gene name, Gene ID and Coordinate (Strand).
2. Peptide information: including the length and sequence of the peptide translated by the translated ncRNA.
3. Sequence and evidence support: including the locus sequence, species, tissue and the experimental methods for the cncRNA.
4. Orthologus of the cncRNA (Human, Chimpanzee, Mouse, Drosophila melanogaster and Zebrafish).
5. References: the information and description from literatures related to the cncRNA.
6. The related subcellular localization, disease and interactions information.
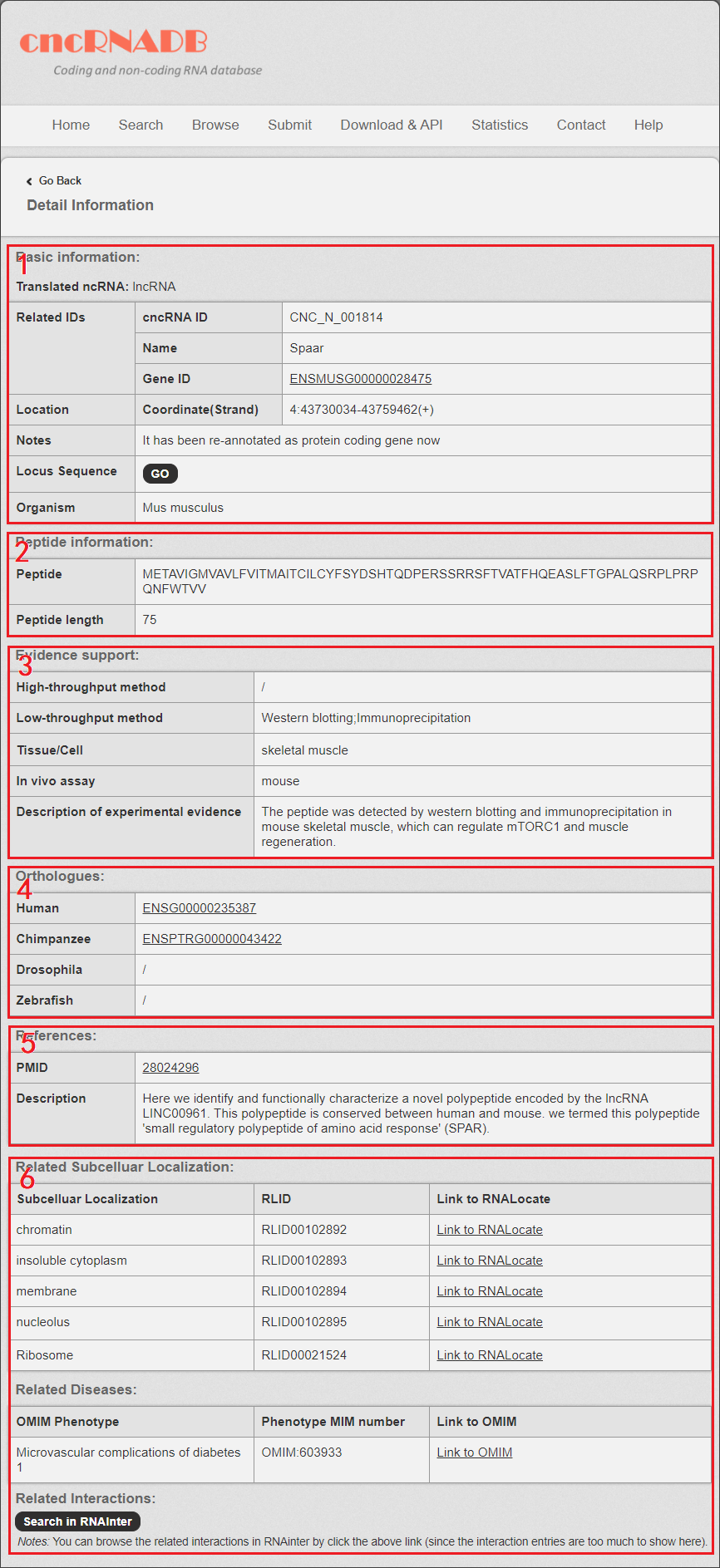
Fig 5-1. Detail page
All the cncRNAs were divided into two categories: translated ncRNAs and untranslated mRNAs. Then the translated ncRNAs was further divided into 6 subcategories: lncRNAs, pri-miRNAs, circRNAs, sRNAs, rRNAs and untranslated regions (UTRs). The untranslated mRNAs were divided into three subcategories: regulatory mRNAs, scaffold mRNAs and sponge mRNAs.
Users can browse translated ncRNA and untranslated mRNA in three different ways, “Browse by RNA type”, “Browse by Organism” and “by Method”. The related cncRNA information is presented by clicking each entry.
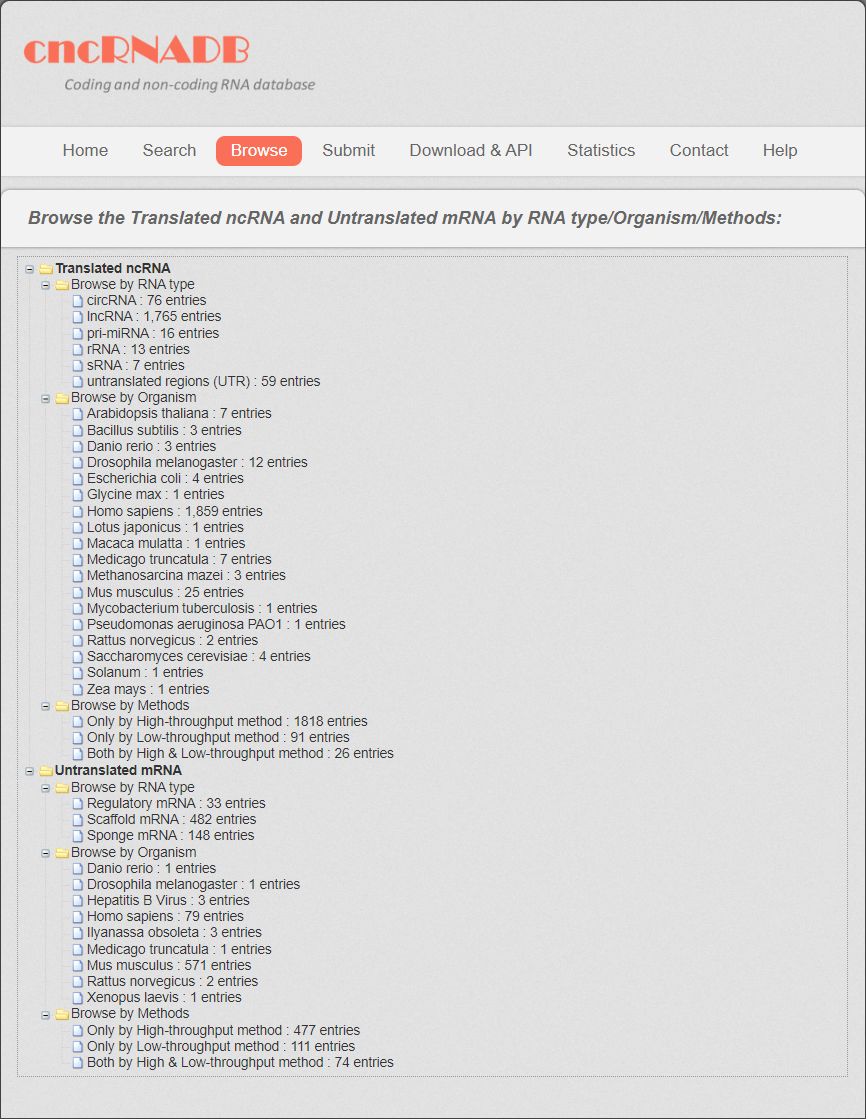
Fig 6-1. Browse page
cncRNAdb provide the download&API page for users. You can download all the cncRNA data in the download&API page.
1. Four tables are provided: table 1: Translated ncRNA, table 2: Scaffold mRNA, table 3: Sponge mRNA, table 4: Regulatory mRNA. The locus sequence is provided as well: locus sequence.fa
2. cncRNAdb provided API for users.
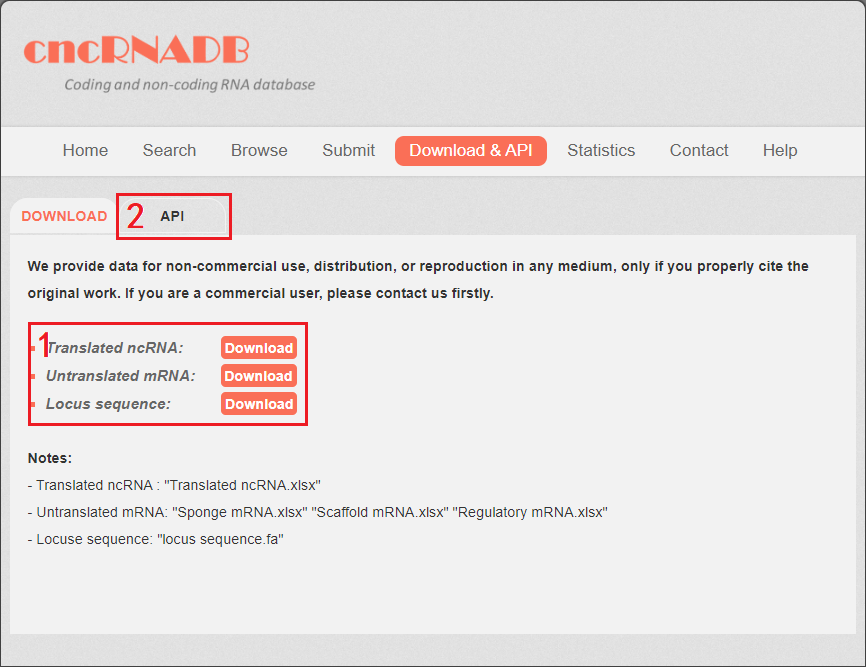
Fig 7-1. Download&API page
