- Source
- Tutorial
- Parameter
- Error Report
Our Relevant Work
- miRDeathDB (Xu J, Li YH et al.,Cell Death Differ. 2012 Sep)
- MNDR (Yuting Wang, Dong Wang, et.al.,Cell Death Disease. 2013.)
ncRNA Related Databases
- miRTarBase (Hsu SD et al.,Nucleic Acids Res. 2011 Jan)
- NPInter (Wu T et al.,Nucleic Acids Res. 2006 Jan:D150-2)
- PRD (Fujimori S et al.,Bioinformation. 2012)
- miRTarBase (Hsu SD et al.,Nucleic Acids Res. 2011 Jan)
- miR2Disease (Jiang Q, Wang Y et al.,Nucleic Acids Res.2009 Jan:D98-104)
- miRNAMap 2.0 (Hsu SD, Chu CH et al.,Nucleic Acids Res.2008 Jan:D165-9)
- human microRNA disease database (HMDD) (Lu M, Zhang Q et al.,PLoS ONE.2008 Oct)
- miRbase (Kozomara A et al.,Nucleic Acids Res. 2011 D152-D157)
- ncRNA Expression Database (NRED) (Dinger ME, Pang KC et al.,Nucleic Acids Res.2009 Jan)
- lncrnadb (Amaral PP et al.,Nucleic Acids Res 39: D146-151.)
- NONCODE (Liu C, Bai B et al.,Nucleic Acids Res.2005 Jan:D112-5)
- Subviral RNA Database (Rocheleau L, Pelchat M et al.,BMC Microbiol. 2006 Mar)
- RNAdb (Bateman A, Agrawal S et al.,RNA. 2011 Nov;17(11):1941-6.)
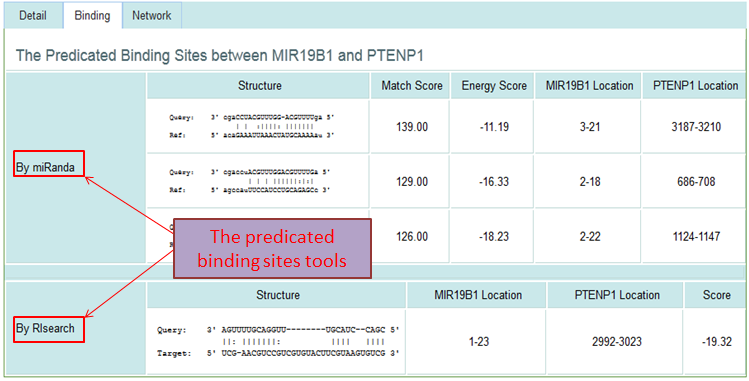
Binding Site Predict Tools
- Protein-RNA Interface Database (PRIDB) (Galperin MY et al.,Nucleic Acids Res. 2007 Jan)
- RBPDB (Cook KB et al.,Nucleic Acids Res. 2011 Jan)
- RsiteDB (Shulman-Peleg A.,Nucleic Acids Research, Vol 37, pp. D369-73)
- bindN (Wang, L. and Brown, S.J et.al., Nucleic Acids Res.)
- miranda (John B et al.,PLoS Biol. 2005 Jul)
- RNAplex (Hakim Tafer and Ivo L. Hofacker.,bioinformatics.2008)
- RNAbindR (Terribilini M et al.,Nucleic Acids Research. 2007 Jul)
Other Gene/Protein Sources
- HGNC (Gray KA et al., Nucleic Acids Res. 2013 Jan 1;41)
- UniProt (The UniProt Consortium.,Nucleic Acids Res. 41: D43-D47 (2013))
- HPRD(Keshava Prasad TS et al.,.Nucleic Acids Res. 2009 Jan)
- Pathway Commons (Cerami et al.,Nucl. Acids Res.2010)
- RNApathwaysDB (Milanowska K et al.,Nucleic Acids Res.2013 Jan;41(Database issue):D268-72.)
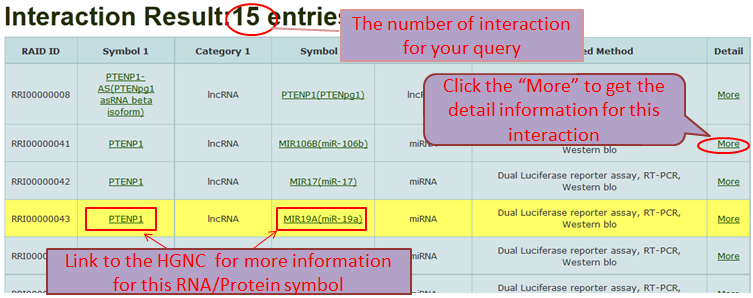
Search RAID by four paths: Keyword, RNA/Protein Category, RNA/Protein Symbol and Validated Method
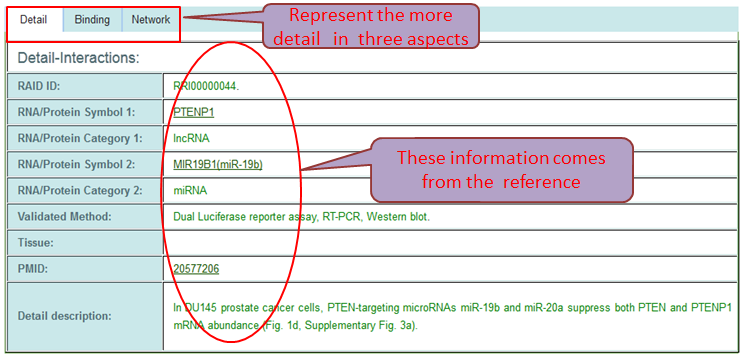
Search Results and Detail Information
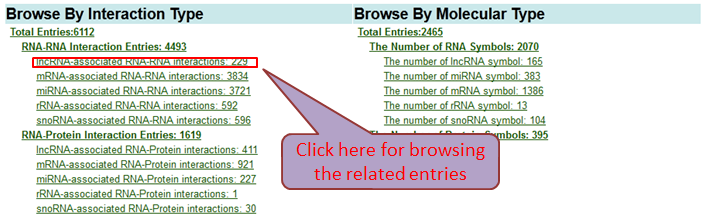
Browse By Interaction Type And Molecular Category
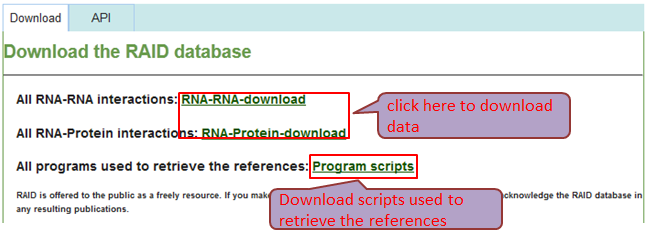
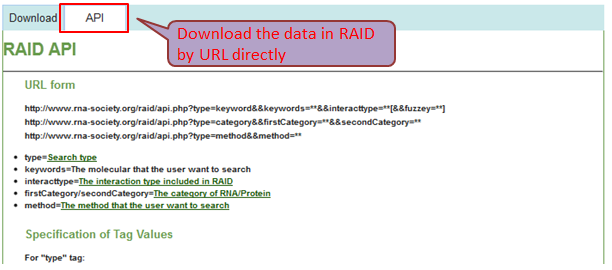
Download & API
Help
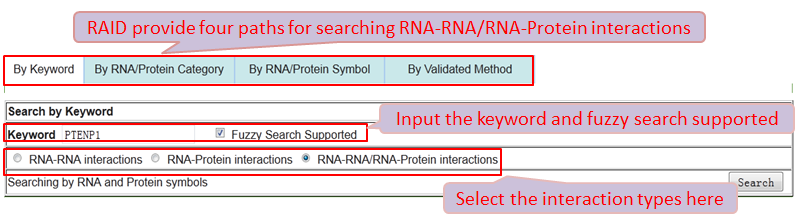
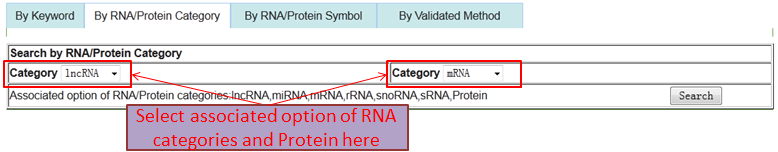
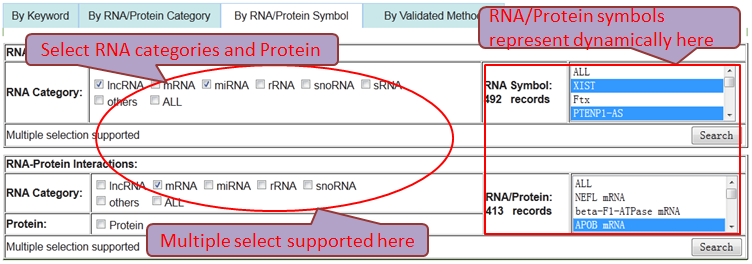
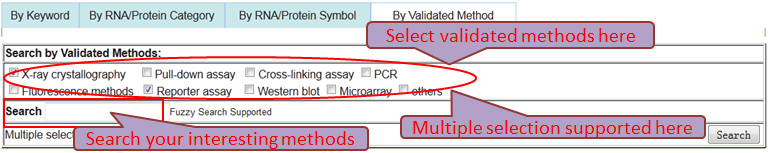
Users can browse and obtain any RNA-associated (RNA-RNA/RNA-Protein) interaction through four paths.
Path 1 (By Keyword): browse the RNA-associated interactions by inputting the keywords(any RNA and Protein Symbol) and fuzzy search supported.
Path 2 (By RNA/Protein Category): represent all RNA-associated interactions by associated option of two different RNA/Protein categories.
Path 3 (By RNA/Protein Symbol): represent the accurate search results by associated option of RNA categories and single RNA/Protein symbol.
Path 4 (By Validated Method): browse the RNA-associated interactions by Validated Methods and multiple selection supported.
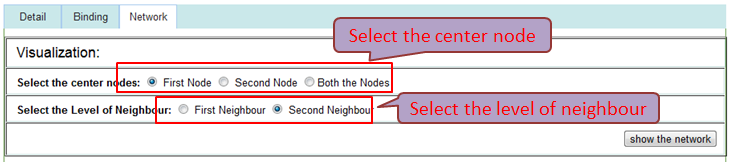
The 'First Node' or 'Second Node' option represents the sub-network of interacting RNA/Protein with the first or second interaction node, the 'Both the Nodes' option represents the sub-network of interacting RNA/Protein with both of interaction nodes.
The 'First Neighbour' represents the sub-network of direct interacting with the center node, the 'Second Neighbour' represents the sub-network of direct or second step interacting with the center node.
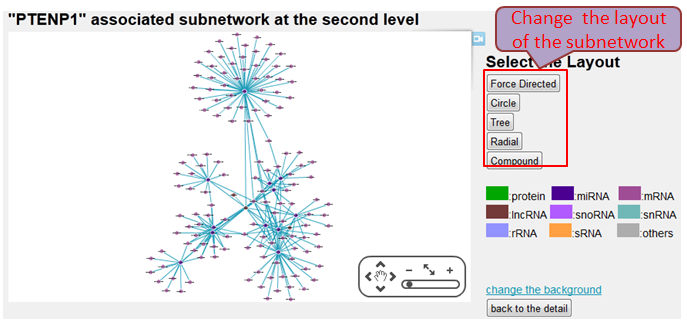
Interaction sub-network based on the two nodes of this interaction may help the researchers represents all interacting partners immediately.

bindN
Web Server: Submit the protein sequences to bindN and get the binding locationhttp://bioinfo.ggc.org/bindn/
Parameter setting
bindN+
Web Server: Submit the protein sequences to bindN+ and get the binding locationhttp://bioinfo.ggc.org/bindn+/
Parameter setting
Miranda
Manual Location: The manual and the parameter of Mirandahttp://cbio.mskcc.org/microrna_data/manual.html
Parameter setting
Pprint
Web Server: Submit the protein sequences to Pprint and get the binding location
http://www.imtech.res.in/raghava/pprint/submit.html
Parameter setting
RIsearch
Manual Location: The manual and the parameter of RIsearch
http://rth.dk/resources/risearch/
Parameter setting
RNABindR
Web Server: Submit the protein sequences to RNABindR and get the binding locationhttp://einstein.cs.iastate.edu/RNABindR/
Parameter setting